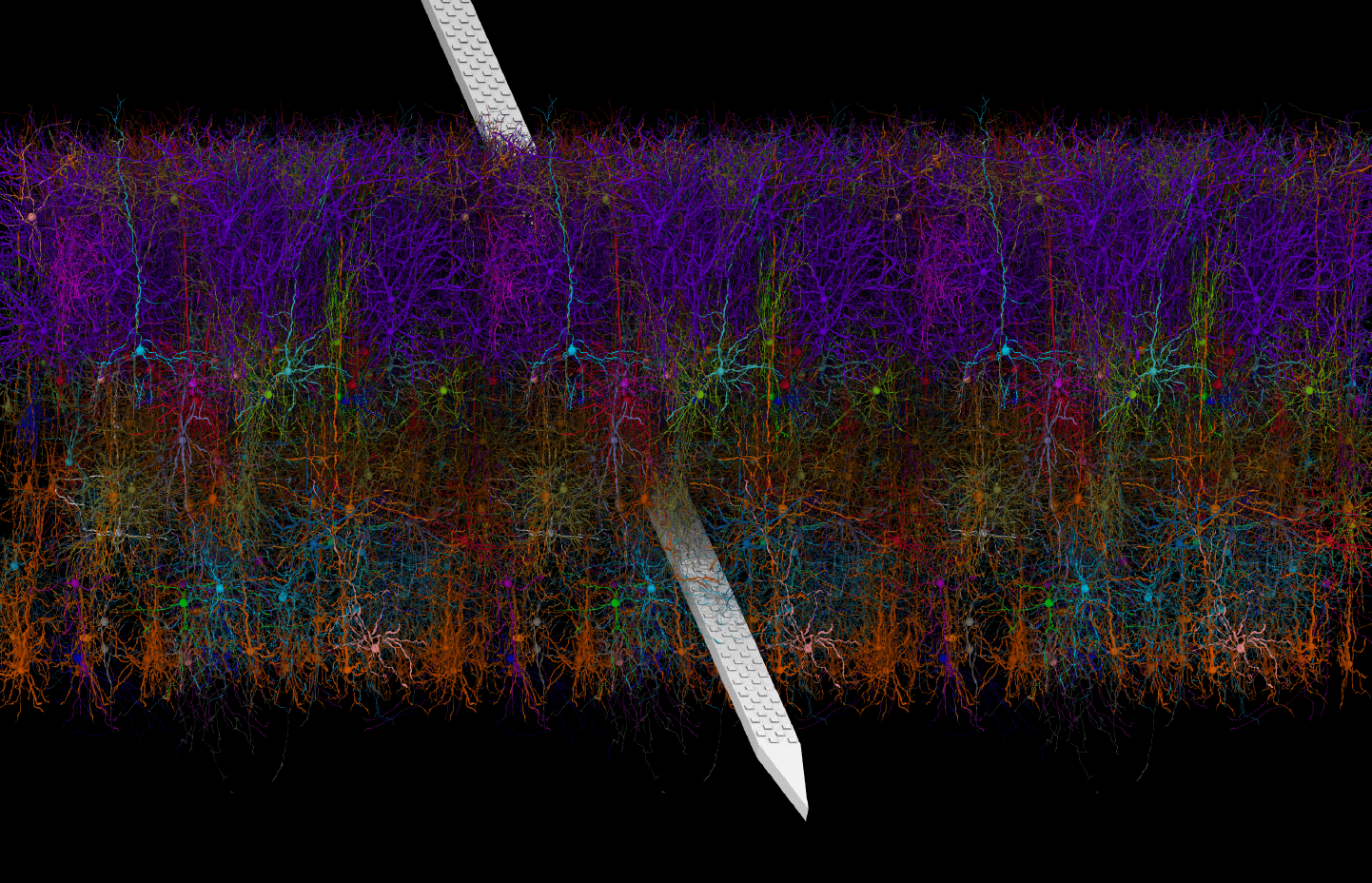
The Allen Institute has developed a variety of single neuron and neural network models of the mammalian neocortex along with modeling and visualization tools for their use. This page summarizes these resources and how to use them.
Biologically realistic models of the mouse primary visual cortex (V1), constructed at biophysically detailed and point-neuron levels.
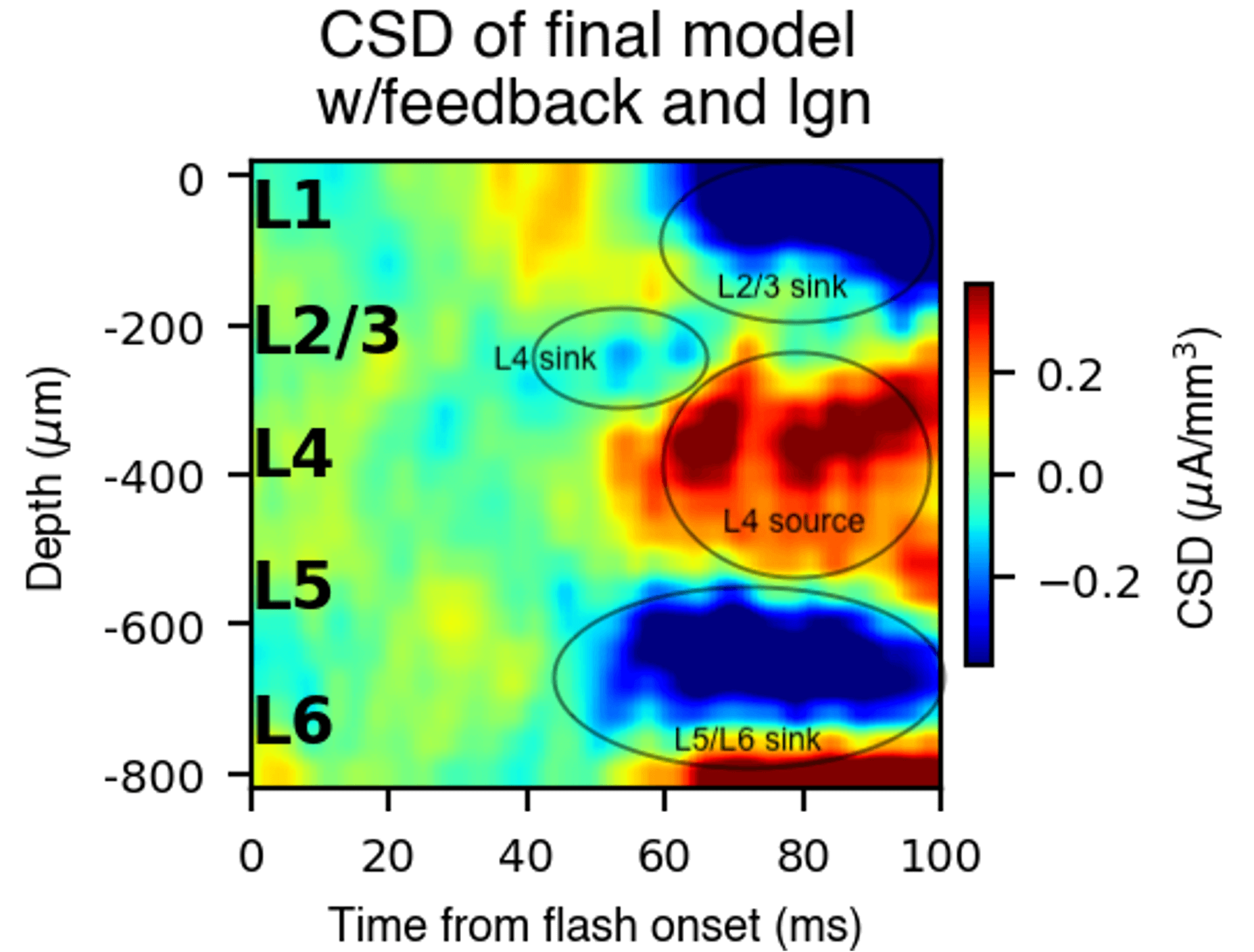
The mouse V1 model was used to simulate electrical signals (LFP/CSD). Feedback from a higher cortical area was added to obtain a better match with LFP recorded in vivo.
Biologically realistic models of the cortical lamina (Layer IV; L4) of mouse primary visual cortex (V1) were constructed at the biophysically detailed and point-neuron levels, to explore mechanisms underlying cortical computations in extensive simulations with a battery of visual stimuli.

MCModels is a Python package providing mesoscale connectivity models for the whole adult mouse brain.
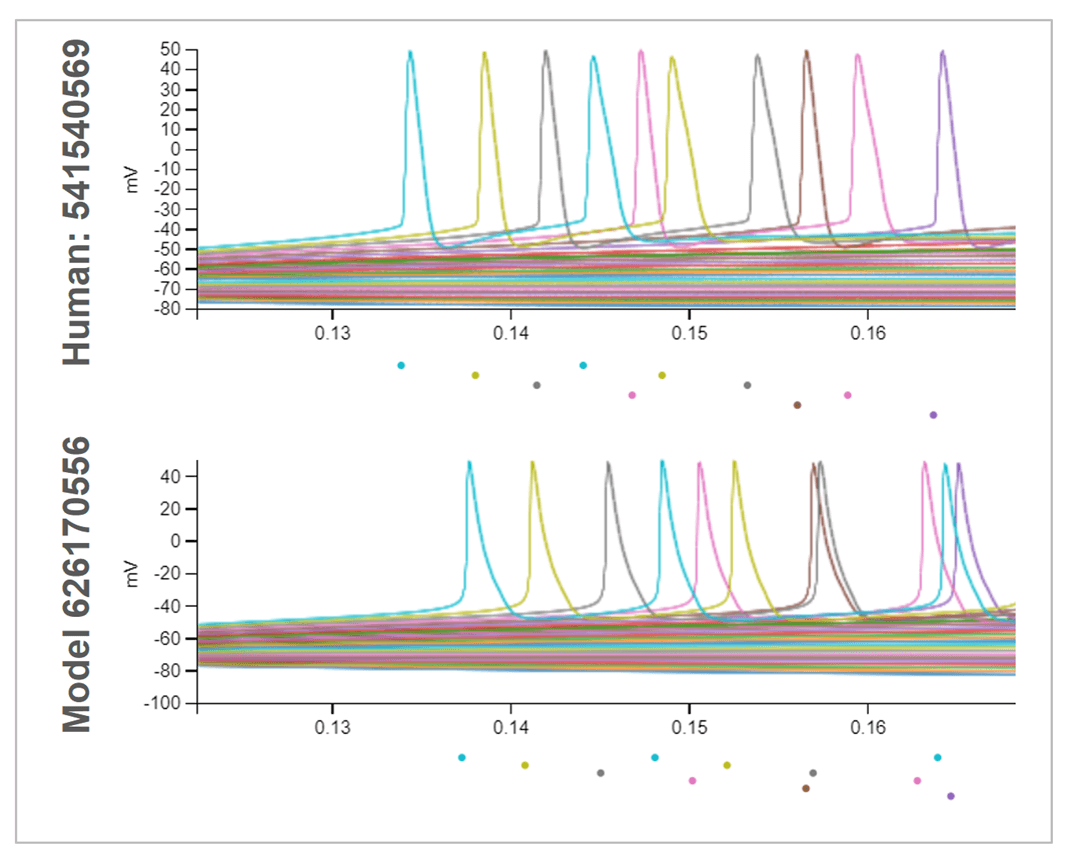
Biophysically-detailed perisomatic-active models are optimized to reproduce the intrinsic firing patterns and action potential properties of individual cells using electrophysiological recordings and morphological reconstructions.
Generalized leaky integrate and fire (GLIF) point-neuron models aim to reproduce the spike times of electrophysiological current clamp data collected from mouse and human cortical neurons.
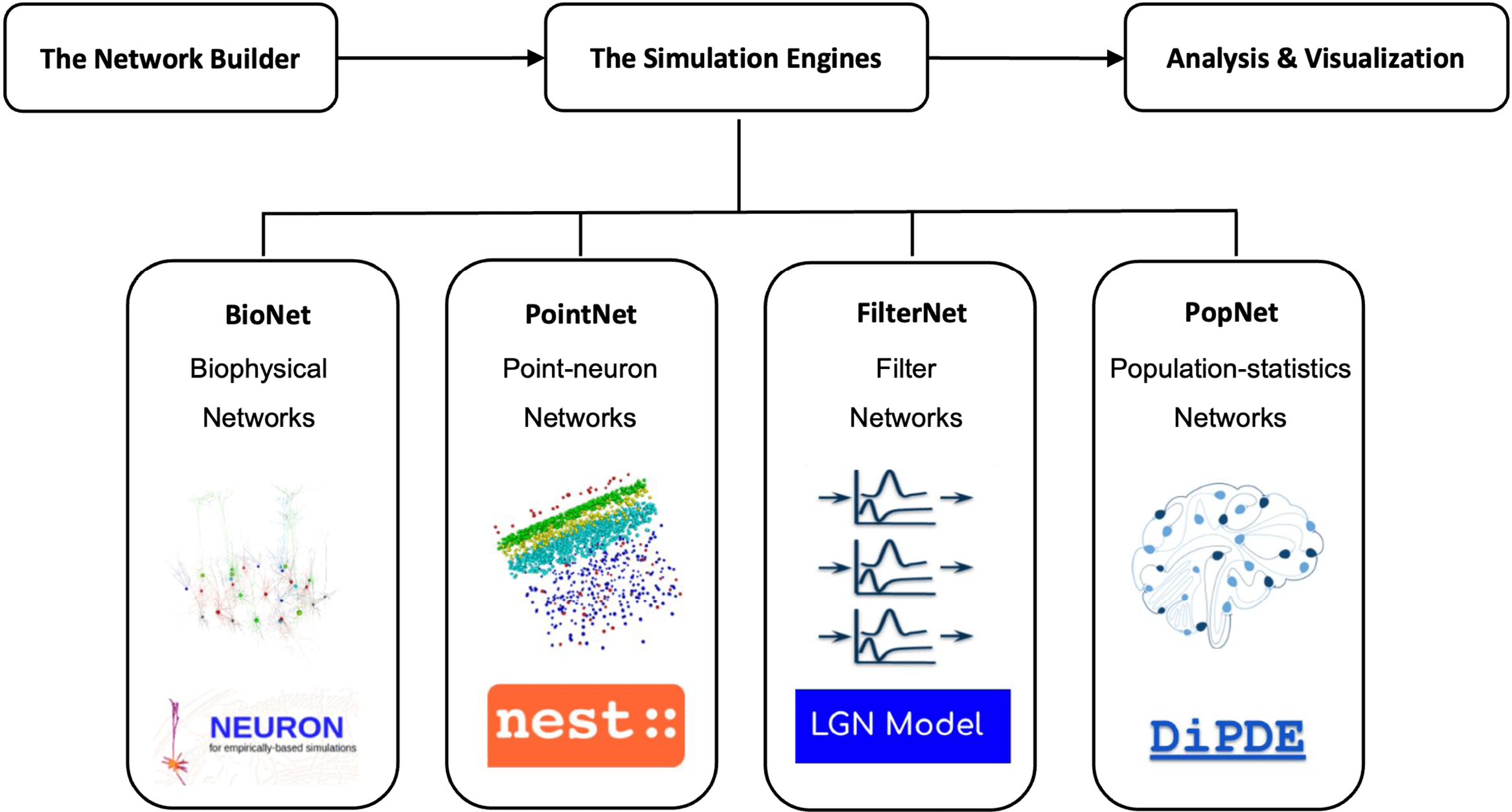
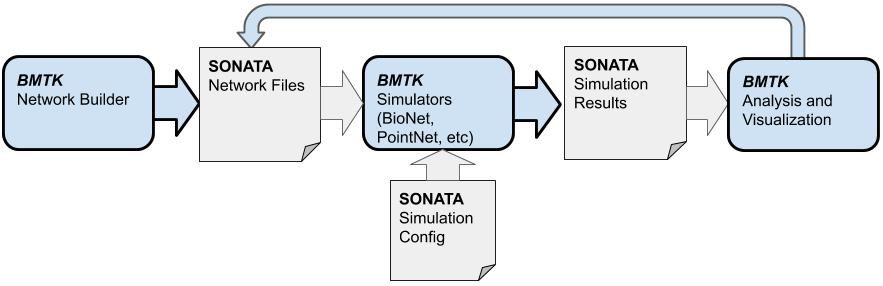
A cross-platform data format for storing large-scale network models and simulation results.

The displacement integro-partial differential equation (DiPDE) population density model refers to a simulation platform for numerically solving the time evolution of coupled networks of neuronal populations. Instead of solving the subthreshold dynamics of individual model leaky-integrate-and-fire (LIF) neurons, DiPDE models the voltage distribution of a population of neurons with a single population density equation. In this way, DiPDE can facilitate the fast exploration of mesoscale (population-level) network topologies, where large populations of neurons are treated as homogeneous with random fine-scale connectivity.